Building BioArena: Kat Yenko on Evaluating Scientific AI Agents
by Ari Wagen · Dec 9, 2025
I recently interviewed Kat Yenko, founder of BioArena, a newly announced platform evaluating the abilities of AI models across scientific domains.
At a high-level, BioArena has three main goals:
- Figure what models are good at and what's still missing.
- Build a community where people can learn, grow their skills, enter the AI for science space, and choose high-impact areas for further work.
- Accelerate scientific progress by helping scientists spend more time with accurate simulations and less time wrangling code / doing dead-end wet lab work.
In our conversation, Kat shared her vision for BioArena, what led her to get started on this project, and how she thinks about evaluating the utility of frontier models for real-world science. If you're interested in getting involved, you can reach out to her on X or Linkedin.
Ari Wagen: I'm talking with you, Kat, and you're building BioArena, which is an evidence-based leaderboard for AI models across scientific domains in the mold of LMArena. Can you tell us about your vision for BioArena?
Kat Yenko: Yeah. It was definitely modeled and inspired by LMArena. I think what they're doing is really cool in terms of going beyond static benchmarks.
With BioArena, the bet that we're making is that the number of scientific models released is going to rapidly increase. Given a world where this is true, how do you decide what scientific models should be used? How do you decide what scientific models to use to best solve your problem? This challenge is what we're aiming to solve with BioArena. It'll be a place where researchers join, ask their questions, and vote on which models work best for their use case. Ultimately, this data gets ranked and sorted in a way where scientists can understand what tools are working for their specific niche domain within science.
Ari: When you use the word "model" here, are you referring primarily to language models like GPT and Claude, or are you thinking about scientific models like Chai, Boltz, and AlphaFold?
Kat: Both. When I use "models," I mean both agentic models and tool models. The agentic models are the Claudes, ChatGPTs, and Geminis, and the tool models are basically what you have on Rowan, which varies from the structure prediction models (Boltz, Chai, and AlphaFold) to a bunch of the other tool models like what you have on Rowan.
Ari: That's great. Tell us a little bit about you: what were you doing before this, and why did you decide to start BioArena?
Kat: The trajectory is kind of funny. I've been in the wet lab, specifically in the human gut microbiome domain, for about eight years now. I've been at Kaleido Biosciences and Stanford and Pendulum and Envivo, and I actually came into biology a little last minute. I was mostly inspired by the fact that I was competing in sports a lot, and I realized that whatever I ate really affected my performance on the field and on the track. And so I hopped into the human gut microbiome space because I really cared about nutrition, and I wanted to have a more mechanistic understanding of how nutrition actually affected performance.
I ultimately ended up here because after being in industry and on the bench for so long and—more recently—playing with computational tools and models, I just realized that there were so many inefficiencies on the bench and so much redundant work that there was potential to resolve. Obviously the models are not perfect right now, but I saw a lot of potential for ways that you could make the wet lab researchers' lives a lot easier by using these tools. And that's how I ended up in BioArena.
I'm building BioArena so that we can move away from more of a brute-force approach into something more precise—and hopefully filter out some of the noise in computational tools—so that we can build trust in the scientific community in these tools.
Ari: Nice, what a great journey. Were you ever using models (whether language models or tool models) while you were a bench scientist?
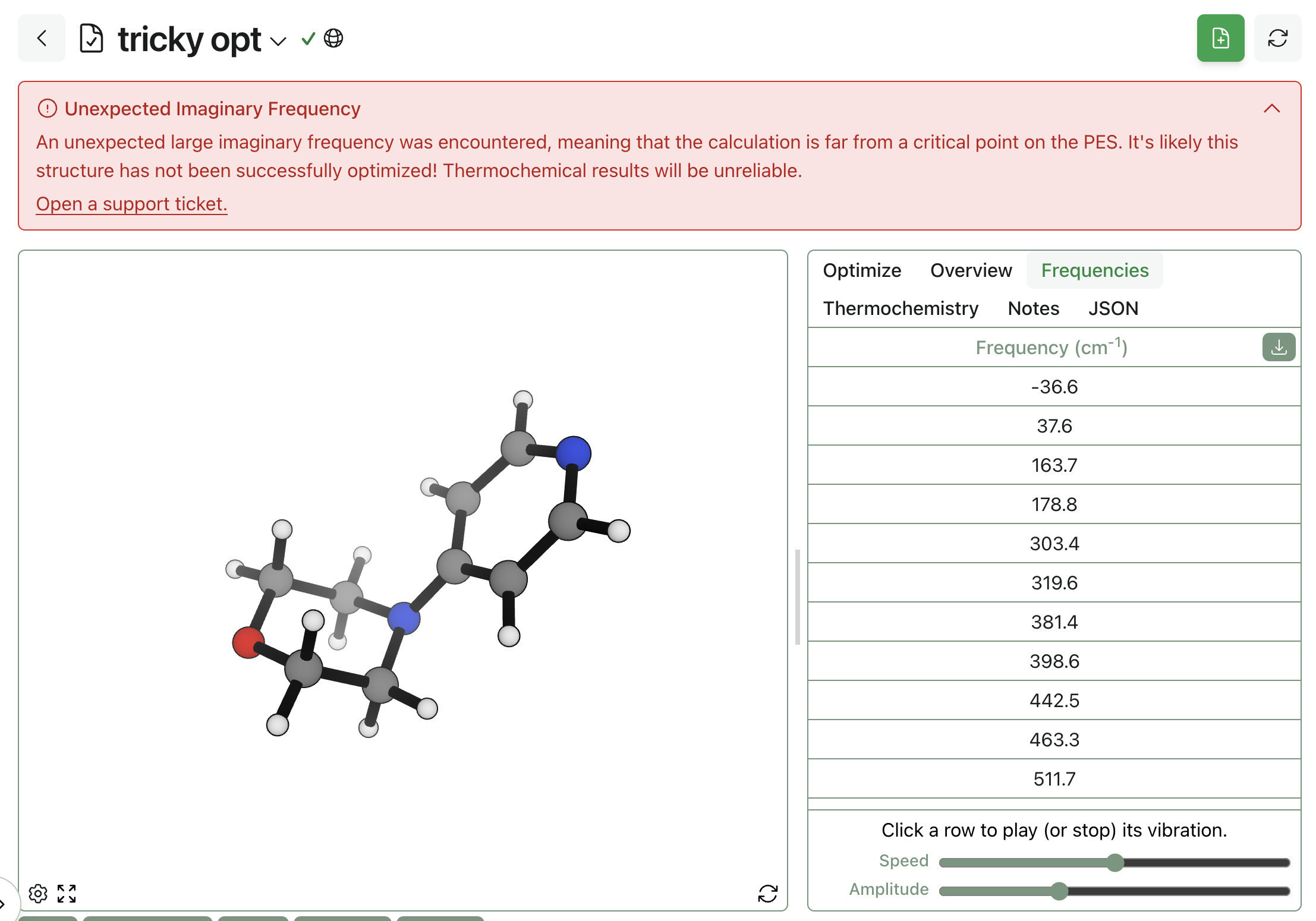
Kat: I started using these tools, mostly language models, about a year and a half ago. I initially started with asking ChatGPT to double-check some basic dilution math calculations, and I wasn't really using any of the tool models. But about a year ago was around the time when I got exposure to Boltz and Chai and AlphaFold. Around that time, at my previous company, we were in the early drug discovery phase and so those fields were somewhat relevant.
I wanted to explore whether you could model a GPCR [G protein-coupled receptor]. And again, my understanding of the models back then was very limited—it still is now—I'm coming from the wet lab world. So I just wanted to understand whether you could model a transmembrane protein and get any signal about whether the complex, unknown compounds in our natural product extracts might bind to it. So to answer your question, I have used the tools before, but never in a way that was actually informing my wet-lab work at the time.
Ari: Yeah. That's really funny. Was it six months ago that you decided to build an MCP server for Rowan?
Kat: Yeah, I think it was about six months ago, in March or April. I found Rowan because in using R and Python when doing typical data analysis for experiments, I was always having trouble with package dependencies and stupid software installations. So when I came across Rowan—which your whole thing is "make it easier for the scientists to do their science"—I found that really intriguing.
I built the MCP because I thought, "Oh, I wonder if you can make it even easier by attaching Claude, which is something that you interact with in natural language, to Rowan, which is more specialized scientific tools, and make it even easier for someone like me back then to just chat with something and have it go use some really specialized scientific tool," because that's not something that is inherently built into Claude or ChatGPT.
Ari: Yeah, a thesis of ours at Rowan is that one reason that these complex domain-specific tools don't get used more is that the experimental scientists don't know enough computation to use the tools that are out there, which are built for computational experts—so they never reach a large enough user base to have the transformative impact that they promise.
Kat: I totally agree with that. I think there is a high barrier to entry for a lot of these wet lab researchers in using these more complex tools.
Ari: I'm curious what you think about the MCP protocol, having built an MCP server and now having done a good bit of testing using an MCP server. Is it a good protocol?
Kat: That's a good question. I'm laughing because I was taking a look at how many tools are shoved in my MCP right now, and it's definitely bloating the context and starting to affect performance on BioArena and the way the agents choose and use tools. So I don't yet know whether it's the most scalable, long-term solution in its current form.
It's still the best option for exposing and orchestrating tools in a standardized way, but I do think that as these systems grow, we'll need something more dynamic. MCP is currently static: every tool gets loaded into context up front, and this doesn't scale when you add dozens of tools. Even with much larger context windows, I don't think appending on more tools in the prompt is a sustainable approach. I think the next step is figuring out how models can discover tools on demand, instead of having the entire tool catalog sitting in context all the time. That would let the system grow without the current trade-offs in performance.
Ari: Yeah, that makes a lot of sense. It seems like a lot of problems are being solved by just throwing more stuff into context, like, "Oh, we're going to give the model memory? Let's condense the past conversation, just add it to context." And that seems like it can scale somewhat, but it's not a very sophisticated solution.
Kat: Yeah, part of the problem with MCP is that every tool has its own description, plus all of its parameters from the Rowan API and a bunch of usage examples that I pulled from the stjames repository on GitHub. So each tool ends up carrying a lot of context. And now that I have around 56 tools in there, a huge portion of what I'm injecting into the model is just tool descriptions.
I think as you start to move towards a space where it's not just one query using one tool and then a response, but it's a more multi-step, chain-of-thought-style process—closer to how a scientist would typically think about problems—the context bloat becomes a bigger issue and just doesn't scale cleanly.
Ari: Yeah. What do you think about multi-agent solutions to this? I've seen some people build agentic systems where there might be a protein-folding agent, a chemical reasoning agent, and a scientific manager agent. What do you think about approaches like that? Do you think that's the future?
Kat: I've played around with a multi-agent type of infrastructure. I think it does help a little bit. I tend to think of each agent like someone on your team—you can have multiple different agents that are each skilled in different niche domains of science, along with a manager agent to coordinate between them. My background is more wet-lab than computational, but I think that makes sense to me. I've moved in that direction, and I think it's working for me right now. I'm not sure yet how well that will translate for everyone.
Ari: Yeah, that makes sense. I want to talk a little bit about your work with LabAgents, which you built, ran, and published the results of after building Rowan MCP but before starting BioArena. What was the goal of LabAgents, and what did you learn?
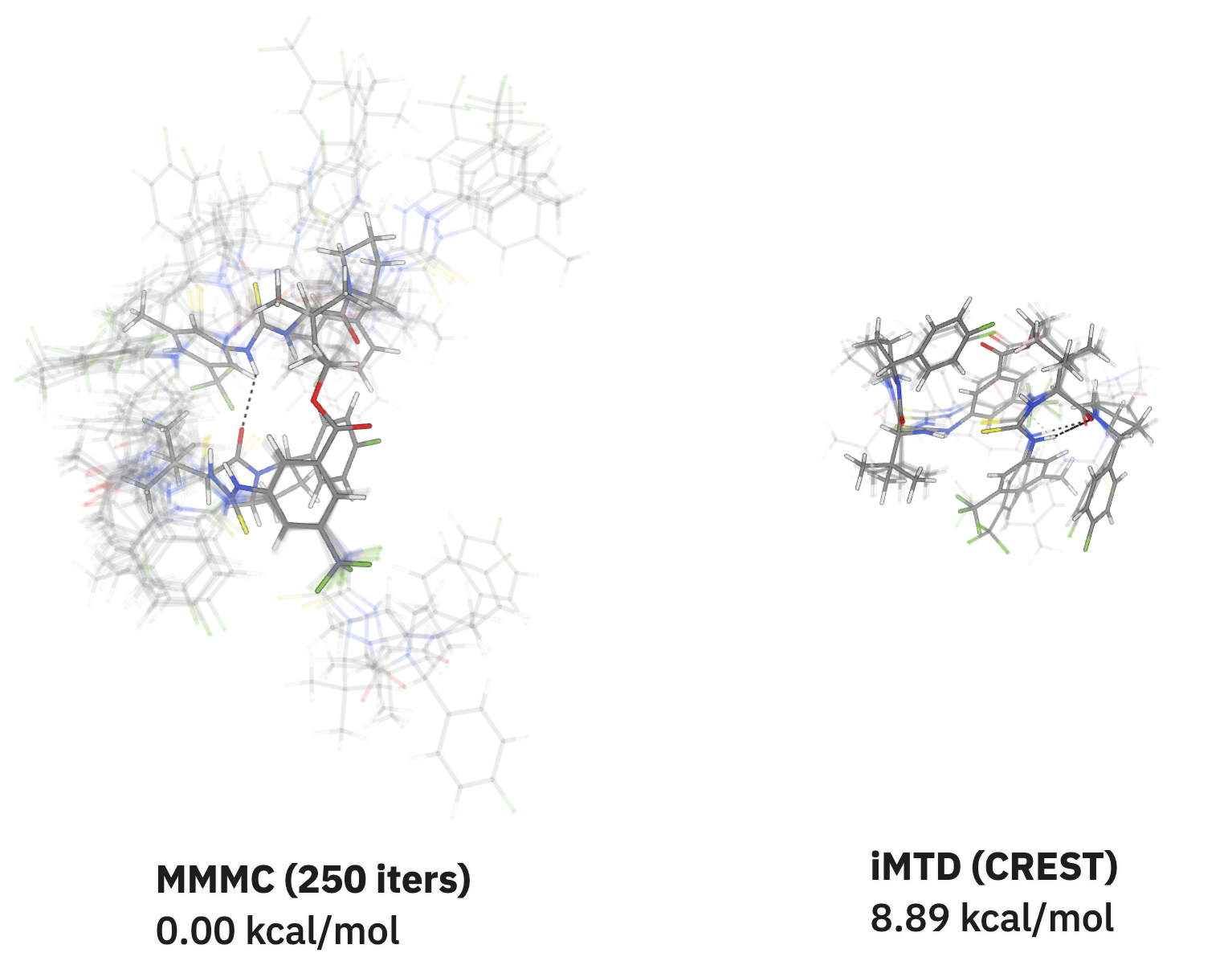
Kat: With Rowan MCP, I built and developed the MCP with Claude. And LabAgents actually came about when Corin had sent me a LinkedIn message, and he sent me Nicholas Runcie's paper. It was about evaluating which models are good at chemical reasoning, and he asked me whether I could evaluate agents specifically for tool use. And that inspired at least my first pass at LabAgents which was, "There's all these models—there's DeepSeek, there's Qwen, there's Claude, there's ChatGPT—can all of them use tools, and do all of them use tools in the same way? If not, how are they different, and is there a way that you can rank them and say, 'Okay, is this agent better at using tools than another one? Is this answer correct or not?'" So that's how LabAgents was born.
I went in super naively, and opened up eight different Cursor windows and had them all run the same set of test questions to evaluate how they answered and how they used the tools that it had via Rowan MCP.
I quickly realized that simple question–answer logs weren't enough, and you had to record reasoning traces to actually understand what was going on. This led to the second iteration of LabAgents, which involved more detailed metrics: what tools each model called, what parameters were used, how many tokens and output tokens they were using, and the full sequence of steps. I went in with some expectations. At the time o3 was considered a really good reasoning model, so I expected it to perform well. I also expected Claude to do reasonably well, partly because I had built the MCP with it, so there was some bias there. But what was surprising was that even though o3 was good at reasoning, it struggled with tool use.
I think the big revelation for me was that different models can reason well but they interact with tools very differently, and that difference is enough to change both the process and final answer.
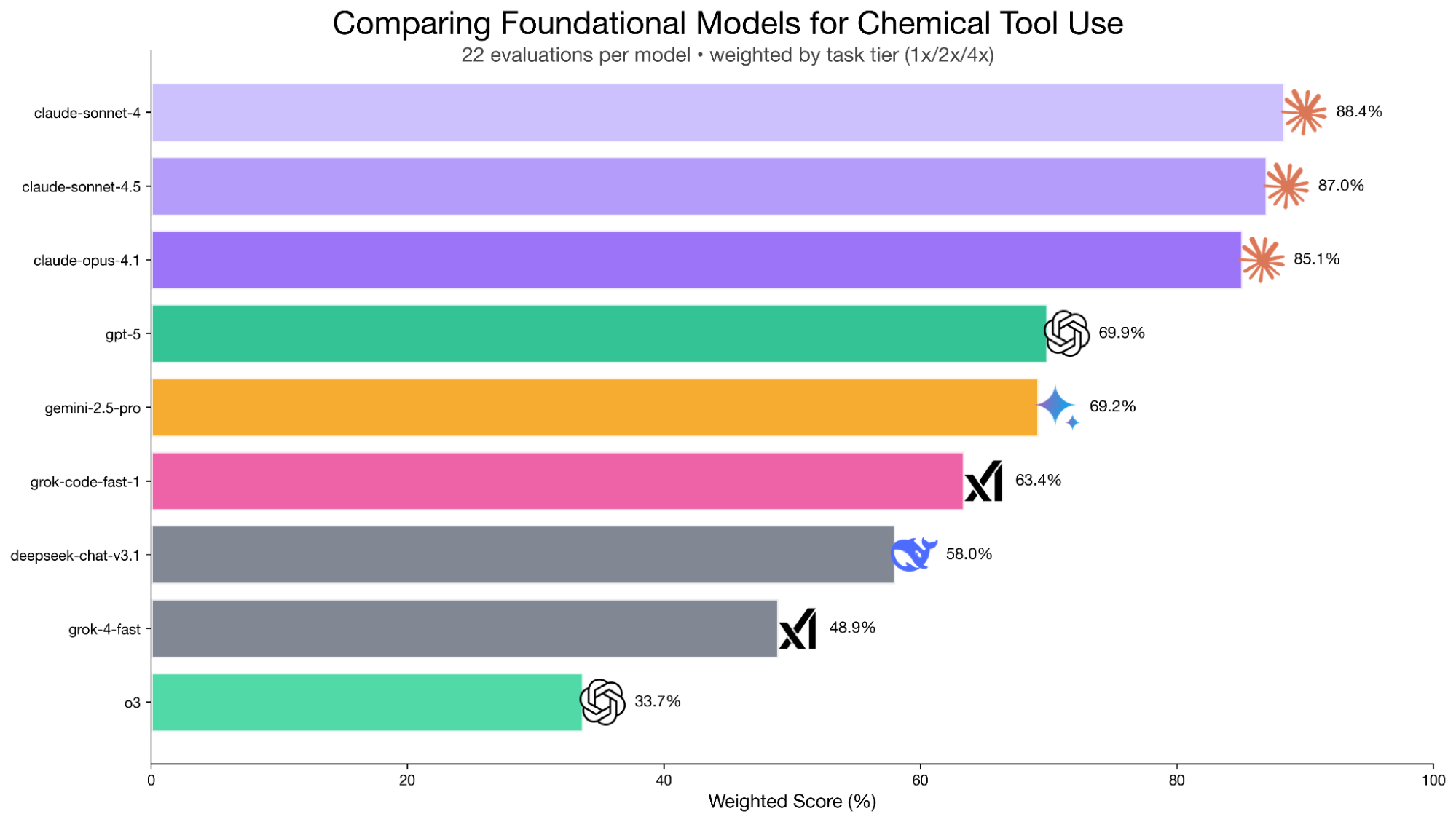
Ari: Yeah. And now you've taken this forward with BioArena. In our conversation so far, we've talked about a few different capabilities of models that we're trying to probe like: "Can these models use tools at all?", "Can they chain tools logically?", "Can they reason about which tools to use and how to work with those outputs?"
I'm wondering, are there other axes that you're hoping to judge or evaluate models on? How do you decompose all these different capabilities? What capabilities do you think the perfect model would have to be a useful co-scientist?
Kat: Yeah, I think beyond just "Is it logical?", "Is the way it uses tools logical?", "Does it come out with the right answer?", at the end of the day the only thing that really matters is: "Is this useful for a scientist?" So no matter what axis you judge every agent or tool on, it just doesn't matter unless it's valuable and useful to the scientist.
Ari: Sure. What's your plan to get scientists involved with evaluating these agents? How are you going to figure out if these agents are useful to scientists?
Kat: So that's the "arena" component in BioArena. Right now, BioArena is hosted on Discord, and it's a place where agents and humans can compete to solve real-world scientific challenges. To take you through how it works, a user submits a question and that initiates a "battle." This battle has two rounds. In the first round, two anonymous agents are pitted against each other to solve this problem using scientific tools, and users can vote on which agent did a better job.
But in the case where the user thinks that they can do better than both of them, they can choose to challenge in the second round. That's where they can submit their own reasoning and answer. If their solution is stronger, they're rewarded for contributing high-quality reasoning and a solid scientific response. I think we can get scientists involved by making sure the reward is enough incentive to get them interested in solving these problems.
Ari: Yeah, I think this is a super cool format, and it opens up so many questions about whether you can teach the models with the human responses. It has the makings of a great AI business.
How do you think about having models evaluate each other versus having humans be the judge? Is it that models aren't ready to choose winners here?
Kat: I think there needs to be both. One piece I left out is that BioArena will also have a validation step that functions as an LLM judge. So it's not that I don't think models are ready to be a judge, but I don't think they should be the only judges.
In BioArena, after an agent or a user submits their answer, it goes through an automated validation step, which is less about making a deep scientific judgment and more about checking that the response falls within the bounds of the question, follows basic constraints, and is coherent. And after this validation check (which is specific to each channel or each domain), it then gets passed to a panel of expert human judges. And at this point they'll be able to see amongst these answers and select between these reasoning traces "Which one is the most logical?", "Which one got here most efficiently?", and "Which answer is the most optimized or the most correct?"
Having a pure LLM-based judge for everything is still really hard. You can set up scoring rules, but if something isn't biologically or chemically reasonable, a human is still much better at catching that nuance. One example that I'm thinking about is the virtual cell challenge that recently happened. Even with good intentions and structured evaluation rules, things can still slip through if they're not grounded in real-world science, and I would trust a human expert more than a model to make that final call right now.
Ari: Yeah, it's a bit circular too. Because I guess a thesis of BioArena is that these models aren't yet perfect at reasoning about biology. So if they can't reason about biology perfectly, then they're not going to be perfect judges. It's a bit like the blind leading the blind if you have the LLMs evaluate the LLMs.
Kat: Yeah, that's why I don't think the validation layer can be purely model-based, even with a strong prompt.
The community input is an important part of grounding the system. Even if you do have a perfect prompt for this validator LLM judge, I think having human input in that feedback loop is what will actually make the judging more trustworthy over time. And at the end of the day you want something that's ultimately helpful for the scientist who is actually doing the work and who's on the bench and using these tools.
Ari: Yeah, that makes a lot of sense. What projects in this area are you following outside of BioArena? What work do you think is cool in AI for science broadly or in agentic science specifically?
Kat: I think the virtual cell is fascinating from the perspective of myself who has worked in the lab and who knows how difficult this stuff is. So I'm really excited by it, but also cautious, because I do think we're far from being able to truly simulate a cell in a way that you could meaningfully run in silico experiments. But that direction itself is really exciting.
I've also been following a lot of the progress in protein structure prediction. Having worked with GPCRs (transmembrane proteins) it's been cool to see tools like AlphaFold, Chai, and Boltz get as good as they have, even though right now they still model proteins in a kind of frozen state. In the long term, I'm excited about seeing GPCRs modeled in context—embedded in membranes and interacting with the whole system rather than existing as isolated, frozen structures.
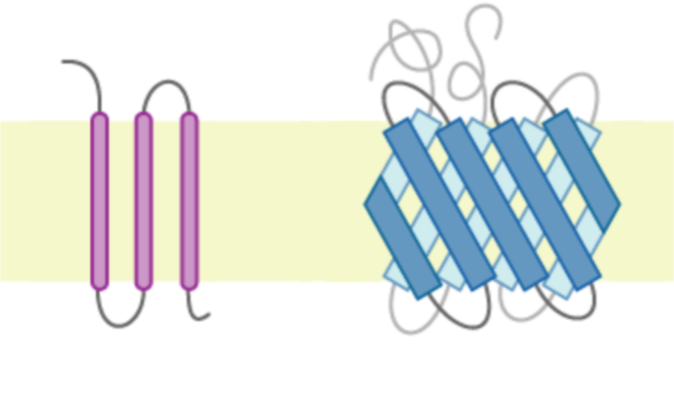
Ari: Totally. And there are a lot of very biologically relevant GPCRs, so it's an area that's very exciting to follow and hopefully we'll see new capabilities come online soon.
Final question: if someone is reading this and they're really interested in BioArena, what should they do?
Kat: Reach out. Whether you're interested in participating in or helping build BioArena, it's just me right now, so I would obviously love input from both the computational side and the wet lab side for what people want to see in the lab. This isn't something that can exist in a vacuum on one side or the other, so if you have ideas, want to contribute, or even just want to experiment with it, I'm open to hearing from you.